Protein-Nucleic Acid Interactions
Protein and nucleic acid interactions are vital to cellular processes. Proteins associate with nucleic acids to mediate transcription and translation of DNA and RNA to decode the information carried by genetic material. In addition, protein–nucleic acid interactions are required to maintain the integrity of DNA and RNA throughout generations. To do so, proteins interact with nucleic acids in processes such as DNA replication, repair and processing, as well as RNA processing and translocation.
Since the development of DNA footprinting in 1978, researchers have been interrogating the composition of protein–nucleic acid complexes. Innovative techniques allow for genome-wide analyses as well as the study of interactions in vivo. Some of the newer methods are outlined below.
Electrophoretic mobility shift assays (EMSA)
A DNA or RNA EMSA is used to determine the specificity of a protein-nucleic acid interaction in vitro. The target nucleic acid is labeled, incubated with potential protein binding partners, then run on a gel. Any protein–nucleic interactions will retard the mobility of the nucleic acid in the gel and result in a shift of the band.
Pull-down assays
Pull-down assays can be performed using either a target nucleic acid sequence or a protein to identify the missing partner(s) in the complex. In this in vitro assay, either the known nucleic acid sequence or the binding protein is attached to an affinity tag, which is used to isolate the complexes from a lysate. If nucleic acid is used for the pull-down, then candidate interacting proteins can be confirmed using antibodies. Alternatively, unknown proteins can be identified using mass spectrometry. If a protein is used for the pull-down, then sequencing can be used to identify any nucleic acids bound to the protein.
Microplate capture and detection
Pull-down assays can be combined with enzyme-linked immunosorbent assays (ELISAs) to perform a technique known as microplate capture and detection. This method is a high-throughput assay to confirm the identity and relative amount of protein partners using antibodies.
ChIP assays
ChIP-seq and ChIP-chip (DNA)
Chromatin immunoprecipitation (ChIP) is used to determine the sequence of DNA bound to a known protein in a living cell.
In a ChiP assay, cells are treated with a crosslinking reagent that stabilizes protein–DNA interactions. An antibody specific for the known protein is then used to purify the protein–DNA complexes. The bound DNA can be identified in several different ways. In traditional ChIP, polymerase chain reaction (PCR) is used to identify the particular DNA sequences bound to the protein. Alternatively, the DNA sequences can be identified using high throughput sequencing in a technique known as ChIP-seq or using a microarray, referred to as ChIP-on-chip.
CLIP-seq (RNA)
Originally termed HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation), cross-linking immunoprecipitation (CLIP)-seq uses UV light as a crosslinking agent to stabilize protein–RNA interactions. The complexes are then immunoprecipitated using an antibody to the target protein. After immunoprecipitation, the complexes are digested with Proteinase K, which removes the bound protein targets and leaves a peptide cross-linked to the protein-binding site. High-throughput sequencing is used to identify the RNA sequences bound by the protein.
CLIP-seq has been used to generate genome-wide splicing factor maps and has been extraordinarily useful in studying microRNAs. Immunoprecipitation of Argonaute, a protein component of the miRNA silencing complex, has been instrumental in identifying miRNA targets, as well as in defining the requirements for miRNA binding.1
Antibody considerations
Immunoprecipitation assays are powerful techniques for studying protein-nucleic acid interactions in vivo. The reliability of the assays is directly dependent on the quality of the antibodies used. Antibodies used for these assays need to be very specific with the high binding efficiency and low background. Only ChIP- or CLIP-verified antibodies should be used.
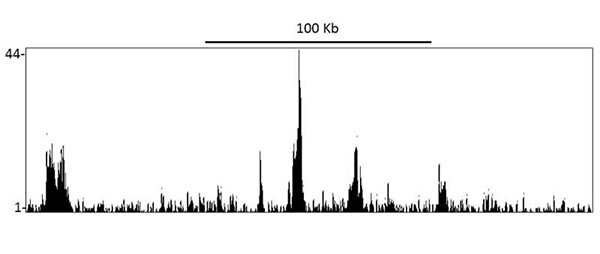
Localization of BRD4 binding sites by in immunoprecipitates from Mia PaCa-2 cells by ChIP-Seq. Antibody: Rabbit anti-BRD4 recombinant monoclonal [BL-149-2H5] (A700-004).
Bethyl Laboratories sells high-quality antibodies for immunoprecipitations that have been used recently to:
- identify the binding sites of a protein involved in transformation resistance.2
- study the interactions of drugs with their target proteins in tumor cells.3
- study gene expression regulation.4
- investigate miRNA processing.5
References
- Chi SW,Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. Jul 23;460(7254):479–486.
- Si H, Scaffidi P, Merchant A, Cam M, Stahlberg E, Mistel T, Fernandez P. 2015. Genome-wise redistribution of BRD4 binding sites in transformation resistant cells. Genomics Data.> Mar 1;3:33–35. [Bethyl antibody used: BRD4]
- Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Lovén J, Sigova A, Smith W, Lee TI, Bradner JE, Young RA. 2014. Genome-wide determination of drug localization. Nat Biotechnol. 32(1):92–6. [Bethyl antibodies used: BRD4, BRD3 (A302-367A, A302-368A), MED1]
- Dardenne E, Espinoza MP, Fattet L, Germann S, Lambert M, Neil H, Zonta E, Mortada H, Gratadou L, Deygas M, et al. 2014. RNA Helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Reports. June 26;7(6):1900–1913. [Bethyl antibodies used: DDX5, hnRNP H1]
- Seong Y, Lim D, Kim A, Seo JH, Lee YS, Song H, Kwon YS. 2014. Global identification of target recognition and cleavage by the Microprocessor in human ES cells. Nucl Acids Res. Nov 10;42(20):12806-12821. [Bethyl antibodies used: DGCR8 (A302-468A, A302-469A)]