Happy Hollidays!
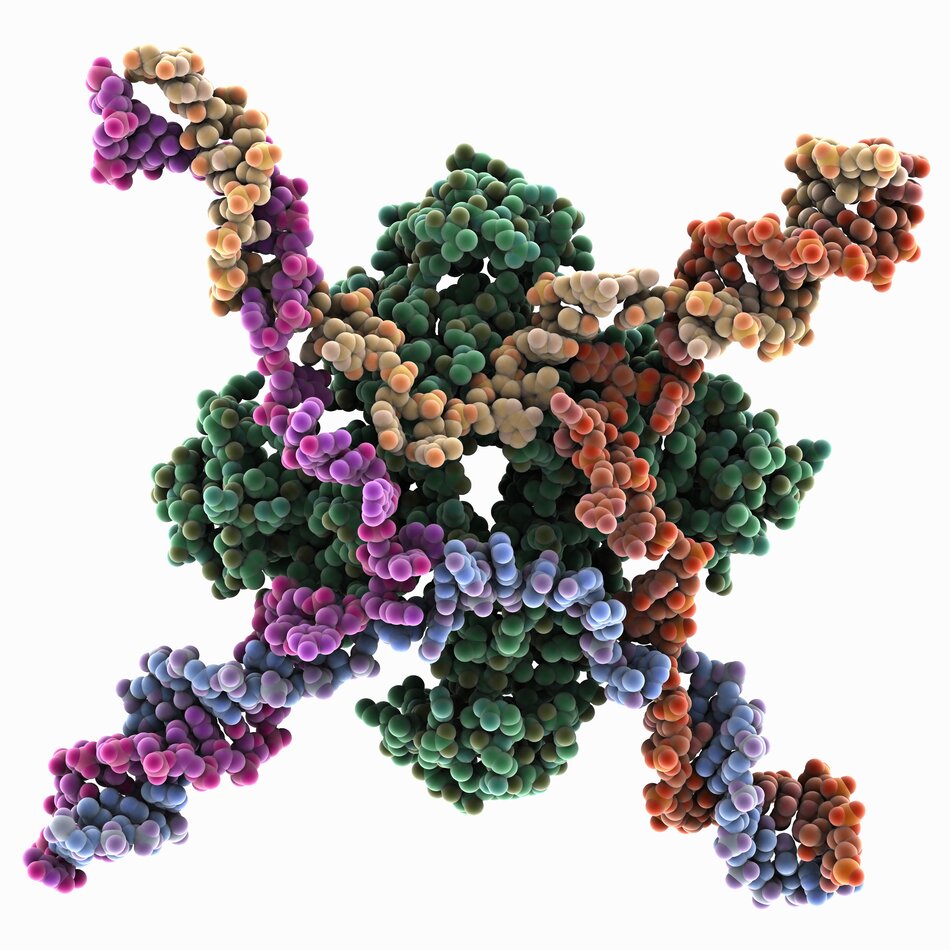
Structure of Holliday Junctions
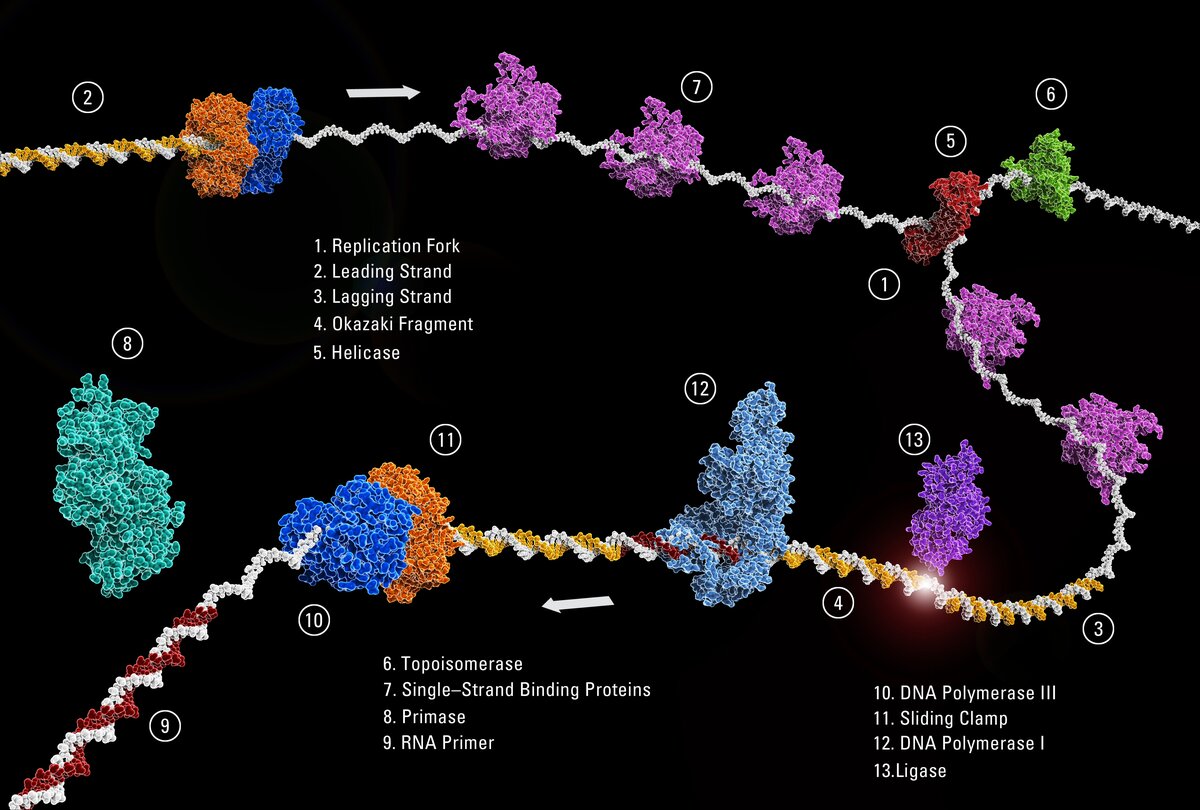
Holliday junctions form a canonical X-stacked structure in the presence of negative ions, and an open-planar structure in the presence of nucleases. The resolution of Holliday junctions to reform a strand of double-stranded DNA can follow multiple pathways. The dissolution pathway involves Bloom’s syndrome helicase (BLM), Topoisomerase IIIα (TopIIIα), Rmi1 (RMI1) and Rmi2 (RMI2) proteins, which primarily dissolve double-Holliday junctions and generate noncrossover products.
The second approach utilizes structure-selective nucleases involved SLX1, SLX4, MUS81 and EME1 proteins to resolve single-Holliday junctions to produce crossover or noncrossover products.
Holliday Junctions in Etiology
Errors in the resolution of Holliday junctions can lead to various genetic disorders, including cancer as well as Bloom’s syndrome, Werner syndrome, and Fanconi Anemia. Bloom's syndrome, a genetic disorder characterized by genomic instability and cancer predisposition, is caused by mutations in the BLM helicase, a key protein involved in Holliday junction dissolution. WRN helicase is involved in multiple DNA repair pathways, including homologous recombination. Defects in WRN can lead to impaired Holliday junction resolution and genomic instability leading to Werner syndrome. The Fanconi Anemia proteins FANCM and SLX4 play a key role in facilitating the migration and resolution of Holliday junctions, which is crucial for proper DNA repair and preventing genome instability associated with the disease.
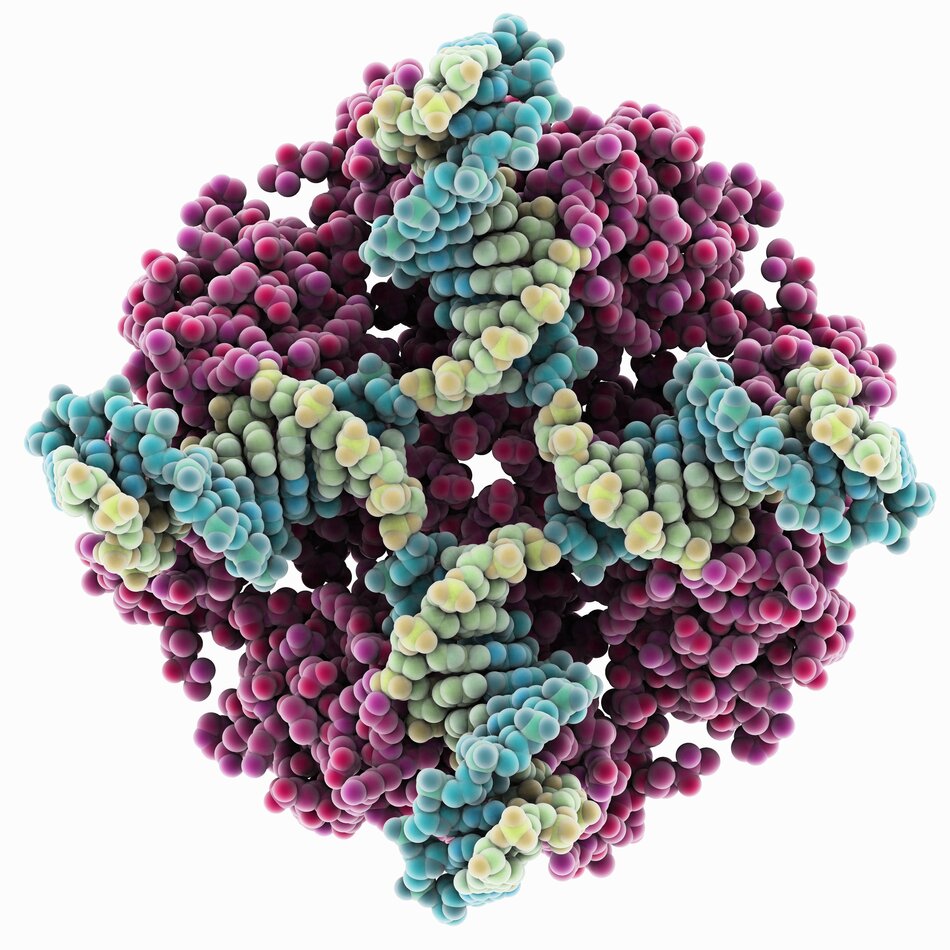
Holliday junction in complex with RuvA in E. coli
Timeline of Holliday Junction Discoveries
- 1964: Holliday junction model proposed in fungal meiosis
- 1973: Holliday junction first visualized in electron microscopy
- 1983: Holliday junction model in DNA double-stranded break
- 2007: First Holliday junction-targeting ligand-based crystal design in vitro
- 2010: Double Holliday junction visualized in DNA double-strand break
- 2014: Overview of bioactivities in human Holliday junction-nucleases in genomic stability
- 2016: Identification and visualization of single-Holliday junction in E. coli
- 2021: Identification of the first bioactive Holliday junction-ligand for oncotherapy
Antibodies Available to Study Holliday Junctions
Timeline adapted from: Song Q, Hu Y, Yin A, Wang H, Yin Q. DNA Holliday Junction: History, Regulation and Bioactivity. International Journal of Molecular Sciences. 2022; 23(17):9730. https://doi.org/10.3390/ijms23179730